AC/DC in the genome: How cells regulate inflammation
Rippe Group | January 16, 2026
Inflammation is finely tuned and deregulated in chronic inflammatory diseases and cancer. Using single-cell genomics and transcriptomics, the group of Karsten Rippe identified ~1,500 genes activated or inhibited during inflammation at different time points. They revealed two complementary regulatory systems, termed AC and DC modules, that finely tune the inflammatory response. Find out more in their publication in Nature Cell Biology.
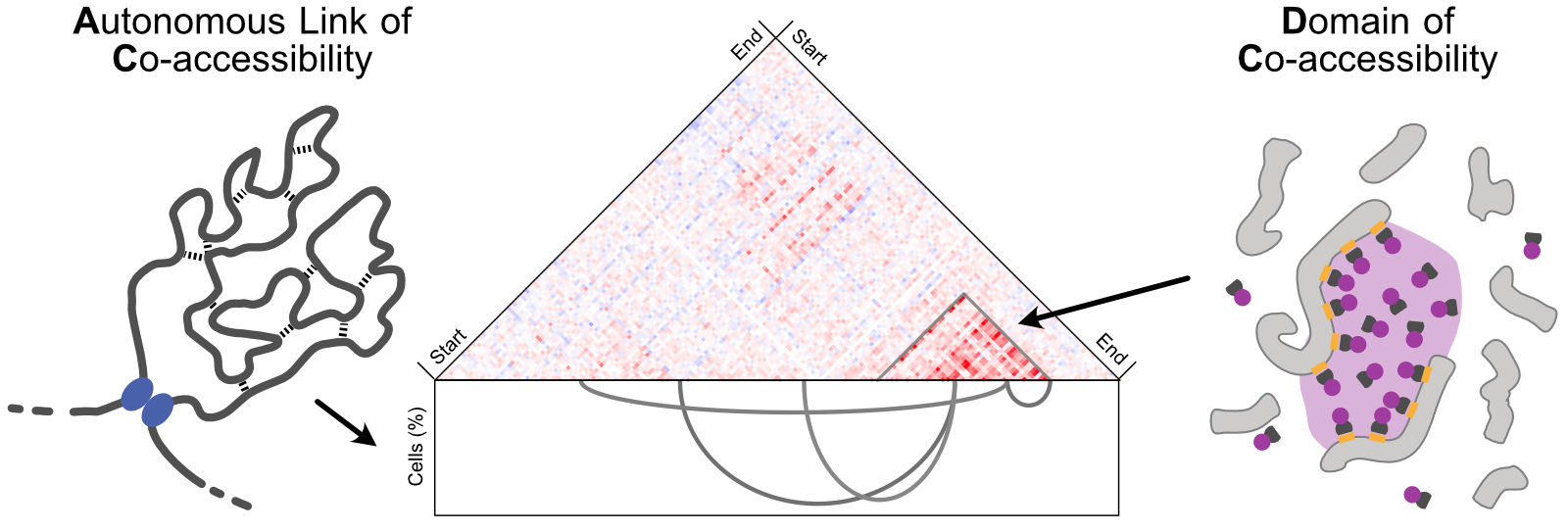
A central regulator of inflammatory gene expression is the signaling pathway induced by tumor necrosis factor (TNF). To understand how cells control the scope, strength, and timing of inflammation, the team of Karsten Rippe, together with their collaborators, studied the response of human endothelial cells to TNF. They developed and applied a new computational method to identify sites in the genome of individual cells that are simultaneously accessible to transcription factor binding. The study uncovered two distinct regulatory systems: AC modules, which are connections between relatively small, separated active genomic sites that frequently control individual genes, particularly those activated later. At the same time, DC modules, which are larger contiguous genomic domains, display a strong local enrichment of transcription factor activity. They can enable rapid, coordinated activation of multiple genes. This dual-layer architecture allows cells to balance speed, strength, and specificity in inflammatory responses. The analysis approach to distinguish AC/DC modules is broadly applicable to other processes, including antiviral responses and leukemia-driving transcription factors.
Publication
Isabelle Seufert, Irene Gerosa, Vassiliki Varamogianni-Mamatsi, Anastasiya Vladimirova, Ezgi Sen, Stefanie Mantz, Anne Rademacher, Sabrina Schumacher, Panagiotis Liakopoulos, Petros Kolovos, Simon Anders, Jan-Philipp Mallm, Argyris Papantonis, Karsten Rippe. Two distinct chromatin modules regulate proinflammatory gene expression. Nat Cell Biol 28, 182–196 (2026), doi: 10.1038/s41556-025-01819-2. Funding was provided by the German Research Foundation (DFG) Priority Program SPP2202.