Biomedical Genomics
Prof. Dr. Carl Herrmann
We study transcriptional control for precise gene expression in cell types and development, using statistical and machine-learning methods to integrate multi-omics data from bulk and single-cell datasets.
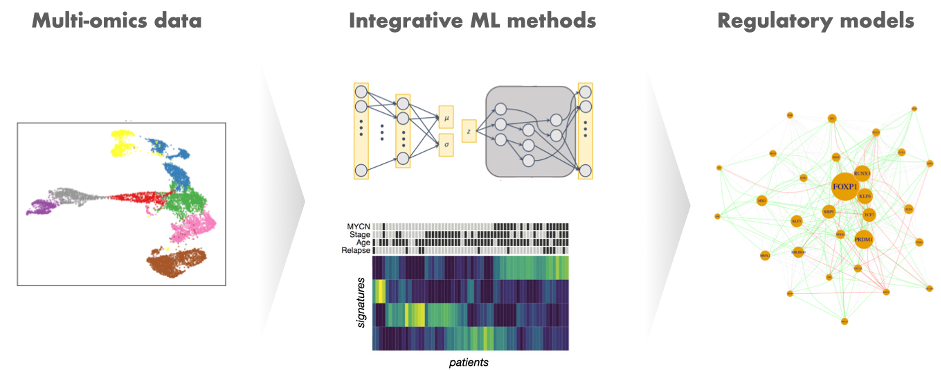
Our group explores how transcriptional programs activate in the right cell type and time, and how this is altered in diseases. Using statistical and machine-learning methods, we integrate multi-omics data from bulk and single-cell datasets to identify molecular signatures and apply transfer learning to describe comorbidity mechanisms. We also work on interpretable deep learning models to relate molecular characteristics with clinical features.
Research Strategy
How is transcriptional control of gene expression regulated in such a way that the right transcriptional programs get activated in the right cell type and the right developmental time? And how is this complex fine tuning altered in diseases, such as cancer, mental disorders and infections? These are the fundamental questions that our group is working on, using a combination of statistical and machine-learning approaches to integrate multiple data types and extract molecular multi-omics signatures representing different regulatory or transcriptional states. We apply this to bulk and single-cell datasets and apply transfer learning strategies to identify shared signatures in different data modalities or diseases, in particular to describe mechanisms of comorbidity. We are increasingly working on interpretable deep learning models (in particular auto-encoders) to provide a fine-grained computational phenotyping of single-cells or patients and relate these molecular characteristics with clinical and macroscopic phenotypic features.

Prof. Dr. Carl Herrmann
- +49 (0)6221 54-51249
- carl.herrmann@bioquant.uni-heidelberg.de
- Find out more information here
Selected Publications
Super enhancers define regulatory subtypes and cell identity in neuroblastoma.
Moritz Gartlgruber, Ashwini Kumar Sharma, Andrés Quintero, Daniel Dreidax, Selina Jansky, Young-Gyu Park, Sina Kreth, Johanna Meder, Daria Doncevic, Paul Saary, Umut H. Toprak, Naveed Ishaque, Elena Afanasyeva, Elisa Wecht, Jan Koster, Rogier Versteeg, Thomas G. P. Grünewald, David T. W. Jones, Stefan M. Pfister, Kai-Oliver Henrich, Johan van Nes, Carl Herrmann, Frank Westermann
Med Image Anal. 2021
Glioblastoma epigenome profiling identifies SOX10 as a master regulator of molecular tumour subtype.
Yonghe Wu, Michael Fletcher, Zuguang Gu, Qi Wang, Barbara Costa, Anna Bertoni, Ka-Hou Man, Magdalena Schlotter, Jörg Felsberg, Jasmin Mangei, Martje Barbus, Ann-Christin Gaupel, Wei Wang, Tobias Weiss, Roland Eils, Michael Weller, Haikun Liu, Guido Reifenberger, Andrey Korshunov, Peter Angel, Peter Lichter, Carl Herrmann & Bernhard Radlwimmer
IEEE Trans Image Process. 2020