Systems Biology of Signal Transduction
Prof. Dr. Ursula Klingmüller
Dynamic pathway models reveal key regulatory mechanisms in cancer progression. Studying erythropoietin receptor (EpoR) signaling, the team identifies cell context-specific information processing, influenced by component abundance. Single-cell level modeling highlights size-dependent heterogeneity in Epo-induced signaling. IFN-induced JAK/STAT pathway dynamics uncover factors affecting pathway responsiveness. TGFβ-induced SMAD complex analysis links relevant trimeric SMAD complexes to target gene expression. Ongoing efforts focus on proteomics and quantification for better predictive modeling of clinical mechanisms.
Research Strategy
Cancer progression is a dynamic process fueled by multiple alterations at the executing level of proteins. To disentangle these complex relations, we develop mechanism- based dynamic pathway models and utilize studies on signal transduction
through the erythropoietin (Epo) receptor, in hepatocytes and in lung cancer cells as paradigms for method and concept development. The Epo/EpoR system enabled us to demonstrate that it is possible to identify key regulatory mechanisms with our dynamic pathway modeling approach, e.g., that the abundance of signaling components determines cell context-specific information processing. We extended this work by employing a mixed effect modelling approach to bridge from the cell population to the single cell level. This allowed us to resolve that the heterogeneity in Epo-induced JAK2/STAT5 signaling in erythroid progenitor cells is primarily caused by processes occurring at cellular or nuclear membranes and is much influenced by the cell size.
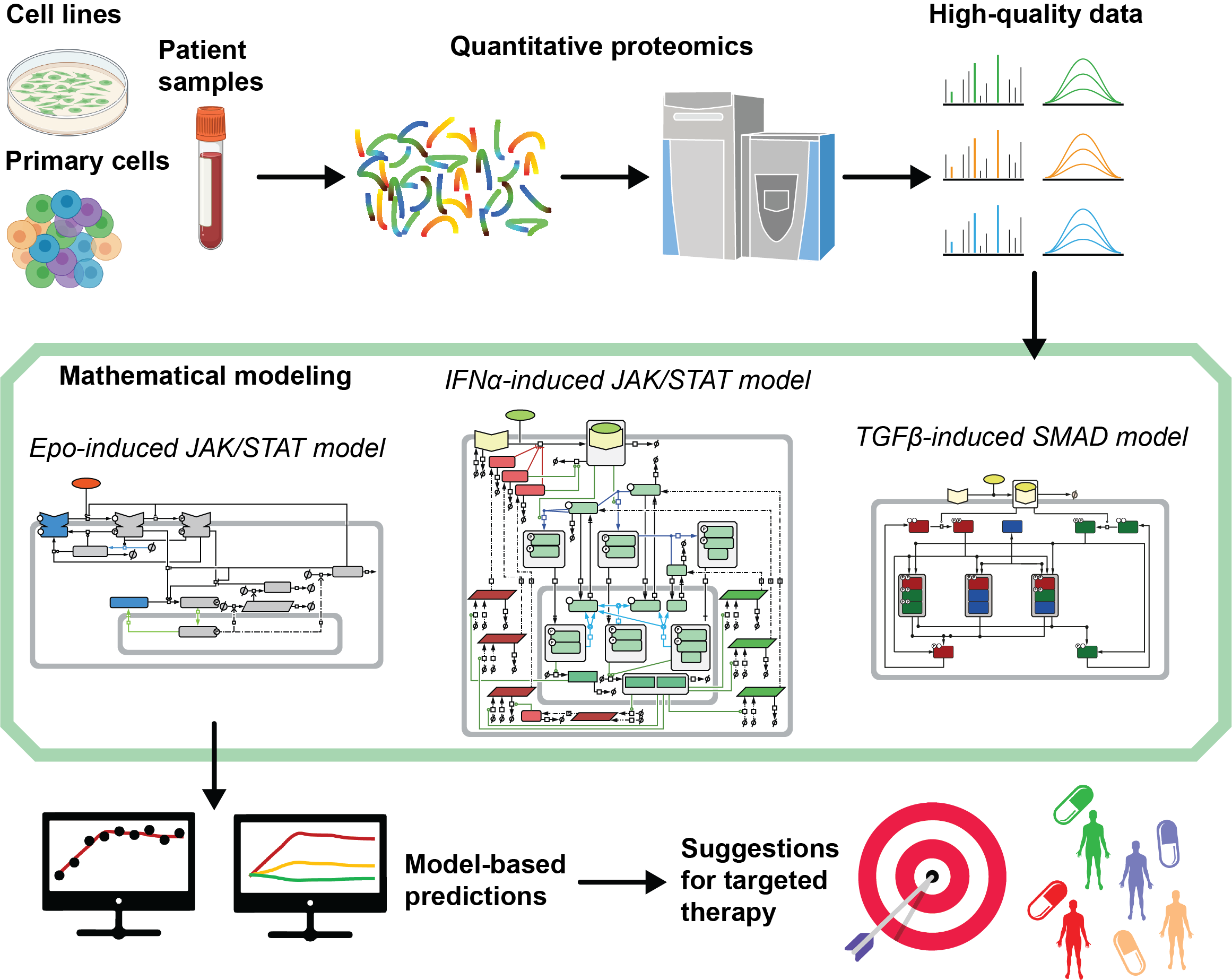
Important components of cellular defense mechanisms are interferons (IFN) that also activate the JAK/STAT signaling pathway. By dynamic pathway modelling, we showed that the timing and the abundance of feedback regulators critically shape the dynamics of pathway activation and provide molecular explanations for pathway desensitization and pathway hypersensitization, depending on the pre-stimulation concentration of IFNα. The quantitative analysis of TGFβ-induced SMAD complex formation in combination with dynamic pathway modelling allowed us to identify among the large number of possible combinations the three most relevant trimeric SMAD complexes and link them to target gene expression. In the coming years, we will continue to invest in quantitative proteomics approaches and develop targeted detection for absolute quantification of all major signaling pathways. The aim is to provide high-quality data from cell lines, primary cells and patient samples for dynamic pathway modelling to resolve mechanism of clinical relevance and improve the predictive accuracy of our modelling approaches.

Prof. Dr. Ursula Klingmüller
- +49 (0)6221 42-4481
- u.klingmueller@dkfz.de
- DKFZ
INF 280
69120 Heidelberg - Deutsches Krebsforschungszentrum (DKFZ)
Selected Publications
More information can be found here
Adlung L, Stapor P, Tönsing C, Schmiester L, Schwarzmüller LE, Postawa L, Wang D, Timmer J, Klingmüller U, Hasenauer J, Schilling M
Cell Rep.2021
Disentangling molecular mechanisms regulating sensitization of interferon alpha signal transduction.
Kok F, Rosenblatt M, Teusel M, Nizharadze T, Gonçalves Magalhães V, Dächert C, Maiwald T, Vlasov A, Wäsch M, Tyufekchieva S, Hoffmann K, Damm G, Seehofer D, Boettler T, Binder M, Timmer J, Schilling M, Klingmüller U
Mol Syst Biol. 2020
Lucarelli P, Schilling M, Kreutz C, Vlasov A, Boehm ME, Iwamoto N, Steiert B, Lattermann S, Wäsch M, Stepath M, Matter MS, Heikenwälder M, Hoffmann K, Deharde D, Damm G, Seehofer D, Muciek M, Gretz N, Lehmann WD, Timmer J, Klingmüller U
Cell Syst. 2018